2. Suggested solutions weekly assignments - lecture 10#
In the lecture notes most functions have been implemented to work for finite elements of any order. However, assemble_mass makes use of
Ae, which is currently only implemented for \(d=\) 1 and 2. Modifyassemble_massto work for arbitrary orders, by adding a functionAdethat returns the element mass matrix of order \(d\). The returned matrix should be a Sympy Matrix including the element lengthh, as used forA1eandA2eabove.def Ade(d=1): ... def assemble_mass(xj, d=1): N = len(xj)-1 Ne = N//d A = np.zeros((N+1, N+1)) Ad = Ade(d) for elem in range(Ne): hj = get_element_length(xj, elem, d=d) s0 = local_to_global_map(elem, d=d) A[s0, s0] += np.array(Ad.subs(h, hj), dtype=float) return A
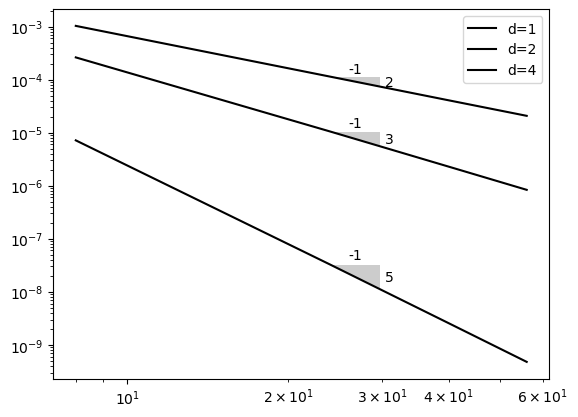
Run the above example with \(u(x) = \exp(\cos(x))\) and show that \(d=4\) leads to a convergence rate of \(N^{-5}\).
Hint
In order to run the example for a range of
Nthat works for both \(d=1, 2\) and \(4\), use \(N \in \{8, 24, 40, 56\}\).
In order to solve this problem and show convergence you can reuse a lot of the code from lecture 10. Only Ade needs to be rewritten in a general manner for any d.
import numpy as np
import sympy as sp
import matplotlib.pyplot as plt
from lagrange import Lagrangebasis
from fem import get_element_length, local_to_global_map, assemble_b, fe_evaluate_v
x, h = sp.symbols('x,h')
def Ade(d=1):
Xj = lambda d: 2*(np.array([sp.Rational(i, d) for i in np.arange(d+1)]))-1 # np.linspace(-1, 1, d+1) only rational
ll = lambda d: Lagrangebasis(Xj(d))
qe = lambda l, r, s: sp.integrate(l[r]*l[s], (x, -1, 1))
A = np.zeros((d+1, d+1), dtype=object)
l = ll(d)
for r in range(d+1):
for s in range(d+1):
A[r, s] = qe(l, r, s)
return (h/2)*sp.Matrix(A)
def assemble_mass(xj, d=1):
N = len(xj)-1
Ne = N//d
A = np.zeros((N+1, N+1))
Ad = Ade(d)
for elem in range(Ne):
hj = get_element_length(xj, elem, d=d)
s0 = local_to_global_map(elem, d=d)
A[s0, s0] += np.array(Ad.subs(h, hj), dtype=float)
return A
Reuse some code from lecture 10:
def assemble(u, N, domain=(-1, 1), d=1, xj=None):
if xj is not None:
mesh = xj
else:
mesh = np.linspace(domain[0], domain[1], N+1)
A = assemble_mass(mesh, d=d)
b = assemble_b(u, mesh, d=d)
return A, b, mesh
Test for d=1
ue = sp.exp(sp.cos(x))
domain = (0, 1)
A, b, xj = assemble(ue, 4, domain=domain, d=1)
uh = np.linalg.inv(A) @ b
yj = np.linspace(domain[0], domain[1], 200)
plt.figure(figsize=(5, 3))
plt.plot(xj, uh, 'b-o', yj, sp.lambdify(x, ue)(yj), 'r--')
plt.legend(['FEM', 'Exact']);
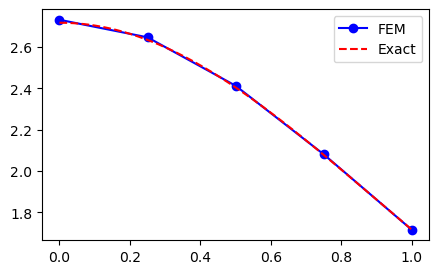
Run L2-error test for a range of meshes and plot the error
def L2_error(uh, ue, xj, d=1, domain=(-1, 1)):
yj = np.linspace(domain[0], domain[1], 4*len(xj))
uhj = fe_evaluate_v(uh, yj, xj, d=d)
uej = sp.lambdify(x, ue)(yj)
return np.sqrt(np.trapz((uhj-uej)**2, dx=yj[1]-yj[0]))
error = {}
NN = (8, 24, 40, 56)
for d in (1, 2, 4):
error[d] = []
for N in NN:
A, b, xj = assemble(ue, N, domain=domain, d=d)
uh = np.linalg.inv(A) @ b
error[d].append(L2_error(uh, ue, xj, d=d, domain=domain))
/var/folders/ql/nly59sfj4dd3spkth6lk0kjw0000gn/T/ipykernel_55654/2891999853.py:5: DeprecationWarning: `trapz` is deprecated. Use `trapezoid` instead, or one of the numerical integration functions in `scipy.integrate`.
return np.sqrt(np.trapz((uhj-uej)**2, dx=yj[1]-yj[0]))
from plotslopes import slope_marker
for d in (1, 2, 4):
plt.loglog(NN, error[d], 'k')
plt.legend(['d=1', 'd=2', 'd=4'])
for d in (1, 2, 4):
slope_marker((24, error[d][1]), (d+1, -1))
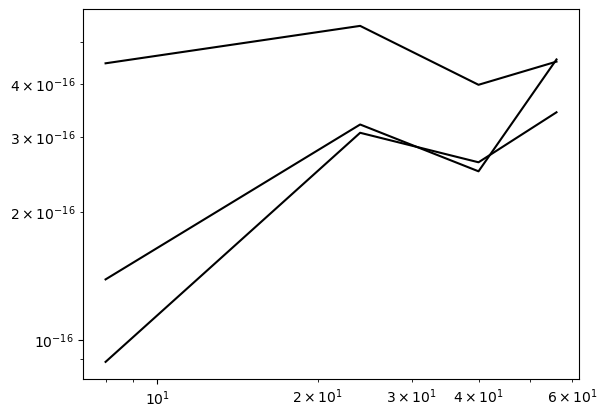
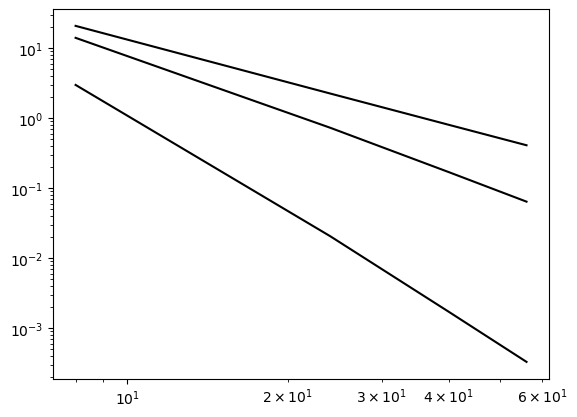
Use FEM to approximate the functions in the weekly assignments from lecture 9. That is,
\(u(x) = |x|, \quad x \in [-1, 1]\)
\(u(x) = \exp(\sin(x)), \quad x \in [0, 2]\)
\(u(x) = x^{10}, \quad x \in [0, 1]\)
\(u(x) = \exp(-(x-0.5)^2) - \exp(-0.25) \quad x \in [0, 1]\)
\(u(x) = J_0(x), \quad x \in [0, 100]\)
All functions can be approximated with the generic implementation already described above, so there is not much new to do. There are some interesting results though. For example, for \(u(x) = |x|\), the results are exact to machine precision for all \(d\).
error = {}
ue = sp.Abs(x)
domain = (-1, 1)
NN = (8, 24, 40, 56)
for d in (1, 2, 4):
error[d] = []
for N in NN:
A, b, xj = assemble(ue, N, domain=domain, d=d)
uh = np.linalg.inv(A) @ b
error[d].append(L2_error(uh, ue, xj, d=d, domain=domain))
for d in (1, 2, 4):
plt.loglog(NN, error[d], 'k')
/var/folders/ql/nly59sfj4dd3spkth6lk0kjw0000gn/T/ipykernel_55654/2891999853.py:5: DeprecationWarning: `trapz` is deprecated. Use `trapezoid` instead, or one of the numerical integration functions in `scipy.integrate`.
return np.sqrt(np.trapz((uhj-uej)**2, dx=yj[1]-yj[0]))
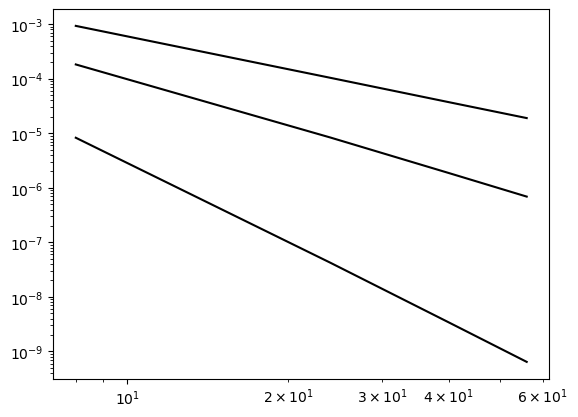
The function \(u(x)=x^{10}\) shows results as expected
error = {}
ue = x**10
domain = (0, 2)
NN = (8, 24, 40, 56)
for d in (1, 2, 4):
error[d] = []
for N in NN:
A, b, xj = assemble(ue, N, domain=domain, d=d)
uh = np.linalg.inv(A) @ b
error[d].append(L2_error(uh, ue, xj, d=d, domain=domain))
for d in (1, 2, 4):
plt.loglog(NN, error[d], 'k')
/var/folders/ql/nly59sfj4dd3spkth6lk0kjw0000gn/T/ipykernel_55654/2891999853.py:5: DeprecationWarning: `trapz` is deprecated. Use `trapezoid` instead, or one of the numerical integration functions in `scipy.integrate`.
return np.sqrt(np.trapz((uhj-uej)**2, dx=yj[1]-yj[0]))
ue = sp.exp(-(x-0.5)**2) - sp.exp(-0.25)
domain = (0, 1)
NN = (8, 24, 40, 56)
for d in (1, 2, 4):
error[d] = []
for N in NN:
A, b, xj = assemble(ue, N, domain=domain, d=d)
uh = np.linalg.inv(A) @ b
error[d].append(L2_error(uh, ue, xj, d=d, domain=domain))
for d in (1, 2, 4):
plt.loglog(NN, error[d], 'k')
/var/folders/ql/nly59sfj4dd3spkth6lk0kjw0000gn/T/ipykernel_55654/2891999853.py:5: DeprecationWarning: `trapz` is deprecated. Use `trapezoid` instead, or one of the numerical integration functions in `scipy.integrate`.
return np.sqrt(np.trapz((uhj-uej)**2, dx=yj[1]-yj[0]))
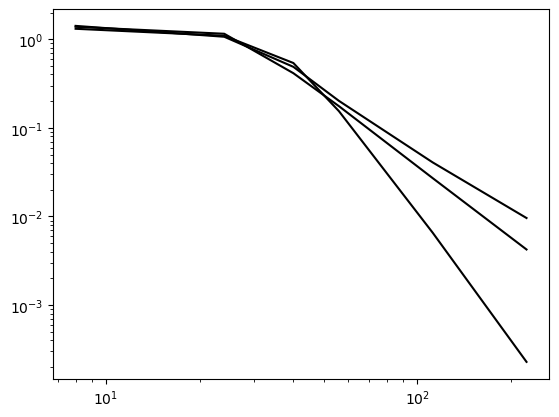
The Bessel function, on the other hand, requires more nodes in order to converge. This is because of the complex shape of the function, that simply cannot be captured with a low number of nodes.
ue = sp.besselj(0, x)
domain = (0, 100)
NN = (8, 24, 40, 56, 112, 224)
for d in (1, 2, 4):
error[d] = []
for N in NN:
A, b, xj = assemble(ue, N, domain=domain, d=d)
uh = np.linalg.inv(A) @ b
error[d].append(L2_error(uh, ue, xj, d=d, domain=domain))
for d in (1, 2, 4):
plt.loglog(NN, error[d], 'k')
/var/folders/ql/nly59sfj4dd3spkth6lk0kjw0000gn/T/ipykernel_55654/2891999853.py:5: DeprecationWarning: `trapz` is deprecated. Use `trapezoid` instead, or one of the numerical integration functions in `scipy.integrate`.
return np.sqrt(np.trapz((uhj-uej)**2, dx=yj[1]-yj[0]))
For the final assignment, simply try to create code like in lecture 10. To get a working version of FEniCS, see this slide.
