Lecture 9 - Function approximation with Chebyshev polynomials and in 2 dimensions#
Short recap#
Legendre polynomials \(\{P_j(x)\}_{j=0}^N, x \in [-1, 1]\) form a basis for the space of all polynomials with order smaller than or equal to \(N\). This space is normally denoted as \(\mathbb{P}_N\). The Galerkin method for approximating \(u(x)\) in the space of polynomials \(\mathbb{P}_N\) would normally be formulated as: find \(u_N \in \mathbb{P}_N\) such that
Here there is no mentioning of the Legendre polynomials as there is no need for them yet. But the Legendre polynomials \(\{P_j\}_{j=0}^N\) may now be chosen as a basis for \(\mathbb{P}_N\) and we can solve Eq. (23) using this basis. The Legendre polynomials are a very good basis because they are orthogonal in \(L^2(-1, 1)\)
and very well suited for approximating other smooth functions. However, the Legendre polynomials are not the only possible basis for \(\mathbb{P}_N\). We can just as easily choose the \(\{x^j\}_{j=0}^N\) and use this basis to solve Eq. (23). But the basis functions \(x^j\) are not orthogonal and the approach is less efficient even though we get exactly the same result. The choice of basis functions is actually very important for the efficiency!
Nodal vs modal basis
The Legendre polynomials are often classified as a modal basis. A modal basis is not connected in any way to mesh points. The Lagrange polynomials, on the other hand, are often referred to as a nodal basis because they rely on mesh points.
We will now introduce yet another modal polynomial basis that is generally considered to be even better than Legendre: The Chebyshev polynomials!
Chebyshev polynomials#
The Chebyshev polynomials can be defined in their most simple and useful form as
for \(x \in [-1, 1]\). Admittedly, this does not look like polynomials at all, it looks like cosine functions! However, an alternative definition is the recursive form
The first 5 polynomials are shown in the figure below
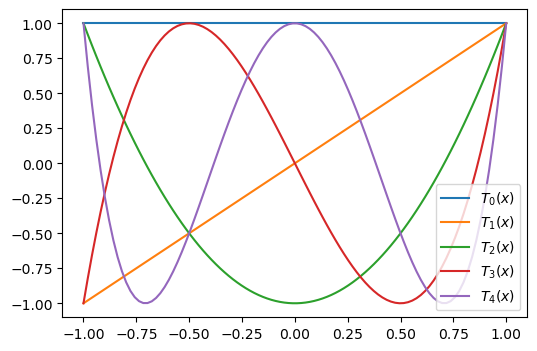
It is noteworthy that all polynomials oscillate between \(-1\) and \(1\). And all polynomials (\(j>0\)) have maximum and minimum values of exactly 1 and -1. All the \(N+1\) extrema points of \(T_N(x)\) are
which are called the Chebyshev points. Another set of points are all the roots of \(T_N(x)\)
The roots are often called the Chebyshev-Gauss points because of their importance in numerical integration through Gaussian quadrature.
Note
Both the roots and the extrema are cosine functions. If we choose to make a change of variables \(x=\cos \theta, \, \theta \in [0, \pi]\), then Eq. (24) becomes
Chebyshev polynomials are very efficient to work with because of the link to the cosine function. The cosine function provides an explicit expression that is easy and efficient to evaluate. The Legendre polynomials, on the other hand, do not have such a nice explicit expression and need to rely on the recursive equation (15) for evaluation.
The Chebyshev polynomials form a basis for \(\mathbb{P}_N\), just like the Legendre polynomials. However, the Chebyshev polynomials are not orthogonal in the \(L^2([-1, 1])\) space! As such, they are not usually used with the Galerkin formulation in Eq. (23).
The Chebyshev polynomials are, on the other hand, orthogonal in a special weighted inner product space. We define the weighted \(L^2_{\omega}(\Omega)\) inner product as
where \(\omega(x)\) is a special weight function. For Chebyshev polynomials defined in \(\Omega = [-1, 1]\) the weight function \(\omega(x) = 1/\sqrt{1-x^2}\). For simplicity we will write the weighted inner product as \((f, g)_{\omega}\).
The Chebyshev polynomials are orthogonal in \(L^2_{\omega}(-1, 1)\) and we can easily get
where \(c_0=2\) and \(c_i = 1\) for \(i > 0\). The weighted squared \(L^2\) norm of Chebyshev polynomials is \(\|T_i\|_{\omega}^2 = \frac{c_i \pi}{2}\).
For Chebyshev polynomials the approximation problem is now solved with the Galerkin method in the \(L^2_{\omega}(-1, 1)\) space: find \(u_N(x) \in \mathbb{P}_N\) such that
Other than the special definition of the inner product, the usage is exactly the same as for the Legendre polynomials. Only the integral is changed.
For a problem where \(x \in [a, b]\) we need to map the true domain to the reference domain, exactly like for the Legendre polynomials. We start with the general definition
which is just a regular Galerkin method using a weighted inner product. Like for Legendre, we use the basis functions in the reference domain, \(\psi_j(x) = T_j(X)\) and the function \(u(x)\) needs a mapping \(u(x(X))\), where \(x(X)\) means that we compute the physical coordinate \(x\) from the reference \(X\). The only new thing is the weight function and this weight function is defined for the reference domain: \(\omega(X(x))\). Naturally, the Chebyshev weight \(\omega(X) = 1/\sqrt{1-X^2}\) only makes sense for the domain \(X \in (-1, 1)\) because the denominator \(1-X^2\) approaches zero for these two edges.
With this in mind we perform a change of variables for (29) where only the function \(u(x(X))\) is written with a mapped argument. The remaining functions are all defined for the reference coordinate \(X\)
As for Legendre the \(dx/dX\) term is constant and can be neglected. We get
Using the orthogonality (27) in (30) we get the Chebyshev expansion coefficients
This should be compared with the Legendre coefficients:
So for both methods it all boils down to one integral.
The weighted inner product requires some extra attention, though. We need to implement the weighted integral
which looks intimidating. However, we now remember the Chebyshev link with the cosine function and change variables with \(X=\cos \theta\)
Furthermore, we can insert for \( 1-\cos^2\theta = \sin^2\theta\) and swap both the direction of the integration and the sign, which leads to
Since \(T_i(\cos \theta) = \cos (i \theta)\) we get the much simpler integral
Using this integral, we get the Chebyshev coefficients
Lets try this with a few examples. First we define the exact weighted inner product
import numpy as np
import sympy as sp
x = sp.Symbol('x', real=True)
k = sp.Symbol('k', integer=True, positive=True)
Tk = lambda k, x: sp.cos(k * sp.acos(x))
cj = lambda j: 2 if j == 0 else 1
def innerw(u, v, domain, ref_domain=(-1, 1)):
A, B = ref_domain
a, b = domain
# map u(x(X)) to use reference coordinate X.
# Note that x is here ref coord.
us = u.subs(x, a + (b-a)*(x-A)/(B-A))
# Change variables x=cos(theta)
us = sp.simplify(us.subs(x, sp.cos(x)), inverse=True)
vs = sp.simplify(v.subs(x, sp.cos(x)), inverse=True)
return sp.integrate(us*vs, (x, 0, sp.pi))
Note the use of the Sympy function simplify with inverse=True, which is required for Sympy to use that \(\cos^{-1}\cos x = x\).
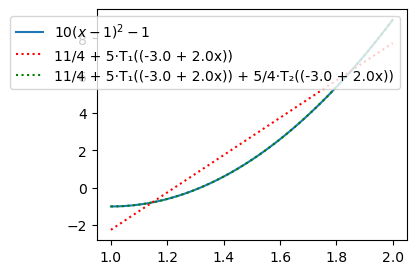
Compute the Chebyshev coefficients for \(u(x) = 10(x-1)^2-1, \, x \in [1, 2]\):
a, b = 1, 2
u = 10*(x-1)**2-1
uhat = lambda u, j: 2 / (cj(j) * sp.pi) * innerw(u, Tk(j, x), (a, b))
Note
The function \(u(x)\) is defined in the true physical domain with \(x \in [a, b]\). Inside the innerw function we map this to \(u(x(X))\)
But we never explicitly create a new symbol X. We just substitute the function \(u\)’s coordinate \(x\) for \(a+(b-a)(x-A)/(B-A)\) and use \(x\) as the reference coordinate.
uhj = uhat(u, 0), uhat(u, 1), uhat(u, 2), uhat(u, 3), uhat(u, 4)
uhj
(11/4, 5, 5/4, 0, 0)
Note that just like for Legendre 3 coefficients are enough to capture the second order polynomial exactly. For plotting we can use the Numpy class Chebyshev with the computed expansion coefficients
from numpy.polynomial import Chebyshev
plt.figure(figsize=(4, 3))
xj = np.linspace(1, 2, 100)
C2 = Chebyshev(uhj[:2], domain=(1, 2))
C3 = Chebyshev(uhj[:3], domain=(1, 2))
plt.plot(xj, sp.lambdify(x, u)(xj))
plt.plot(xj, C2(xj), 'r:')
plt.plot(xj, C3(xj), 'g:')
plt.legend(['$10(x-1)^2-1$', f'{C2}', f'{C3}']);
Lets try the more difficult function
Since the weighted inner product now becomes difficult (at least timeconsuming) for Sympy to integrate exactly, we create a numerical weighted inner product as well.
from scipy.integrate import quad
def innerwn(u, v, domain, ref_domain=(-1, 1)):
A, B = ref_domain
a, b = domain
us = u.subs(x, a + (b-a)*(x-A)/(B-A))
us = sp.simplify(us.subs(x, sp.cos(x)), inverse=True)
vs = sp.simplify(v.subs(x, sp.cos(x)), inverse=True)
return quad(sp.lambdify(x, us*vs), 0, np.pi)[0]
u = sp.exp(sp.cos(x))
#u = 1 / (1+16*x**2)
#uhat = lambda u, j: 2 / (cj(j) * sp.pi) * innerw(u, Tk(j, x), (-1, 1)) # slow
uhatn = lambda u, j: 2 / (cj(j) * np.pi) * innerwn(u, Tk(j, x), (-1, 1))
The function uhatn computes the Chebyshev expansion coefficients corresponding to Eq. (34) and we compute the 25 first coefficients below. For comparison we also compute the 25 first Legendre coefficients, corresponding to Eq. (19).
uc = []
N = 25
for n in range(N):
uc.append(uhatn(u, n))
# For Legendre:
from numpy.polynomial import Legendre
def innern(u, v):
uj = lambda xj: sp.lambdify(x, u)(xj)*v(xj)
return quad(uj, -1, 1)[0]
uj = lambda u, j: (2*j+1) * innern(u, Legendre.basis(j))/2
ul = []
for n in range(N):
ul.append(uj(u, n))
Note
Note the numerical implementation of the Legendre inner product. Here we could have used simply
def innern(u, v):
return quad(sp.lambdify(x, u*v), -1, 1)[0]
uj = lambda u, j: (2*j+1) * innern(u, sp.legendre(j, x))/2
However, this is much more susceptible to roundoff errors. Just try it!
Plot both Chebyshev and Legendre coefficients in a semilogarithmic plot. Since all the odd coefficients are zero for both Chebyshev and Legendre, we plot only the even coefficients. The odd coefficients are zero because \(u(x)\) is an even function. The Chebyshev and Legendre polynomials are all such that \(\psi_{2i}(x)\) are even, whereas \(\psi_{2i+1}(x)\) are odd. See the Chebyshev basis functions.
plt.figure(figsize=(5, 3))
plt.semilogy(np.arange(0, N, 2), uc[::2], '+',
np.arange(0, N, 2), ul[::2], 'ko', fillstyle='none')
plt.legend(['Chebyshev', 'Legendre']);
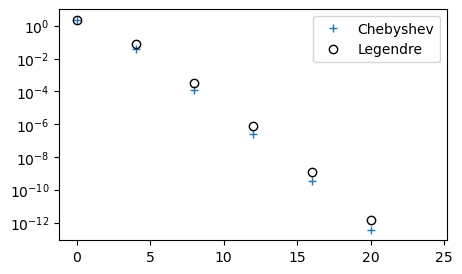
We note that the Chebyshev coefficients are smaller than the Legendre. This is an indication that the Chebyshev polynomials are performing better. However, to say something about the true accuracy we really need to compute the \(L^2\) error norm. Below we compute the \(L^2\) error norm (note, not a weighted norm) for both Legendre and Chebyshev as a function of \(N\).
from numpy.polynomial import Legendre
def L2_error(uh, ue, space=Legendre):
xj = np.linspace(-1, 1, 1000)
uej = sp.lambdify(x, ue)(xj)
err = []
for n in range(0, len(uh), 2):
uj = space(uh[:(n+1)])(xj).astype(float)
err.append(np.sqrt(np.trapz((uj-uej)**2, dx=xj[1]-xj[0])))
return err
errc = L2_error(uc, u, Chebyshev)
errl = L2_error(ul, u, Legendre)
np.save('./err_u', np.array([errc, errl]))
/var/folders/ql/nly59sfj4dd3spkth6lk0kjw0000gn/T/ipykernel_55623/3566146242.py:9: DeprecationWarning: `trapz` is deprecated. Use `trapezoid` instead, or one of the numerical integration functions in `scipy.integrate`.
err.append(np.sqrt(np.trapz((uj-uej)**2, dx=xj[1]-xj[0])))
plt.figure(figsize=(5, 3))
plt.loglog(np.arange(0, N, 2), errc, '+',
np.arange(0, N, 2), errl, 'ko', fillstyle='none')
plt.legend(['Chebyshev', 'Legendre']);
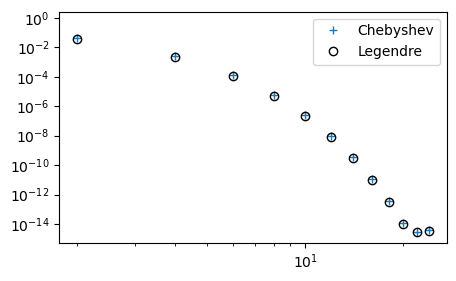
So there is hardly any difference at all in accuracy.
Note
This very fast convergence down to machine precision is often referred to as spectral accuracy. It is actually faster than any finite difference method, which always leads to linear decay in the loglog plot above since the error in the finite difference method can be written as some integer order \(r\), such that the error is \(\mathcal{O}(\Delta x^r)\).
Two-dimensional functions#
We can approximate a two-dimensional function \(u(x, y)\) using a two-dimensional function space \(W\), such that
where \(\Psi_{i}(x, y)\) is a two-dimensional basis function and \(\{\Psi_i(x, y)\}_{i=0}^M\) is a basis for the function space \(W = \text{span}\{\Psi_i(x, y)\}_{i=0}^M\). Just like in one dimensional space we will write \(u_N\) for the series expansion that approximates \(u\).
There are not all that many two-dimensional basis functions and a more common approach is to use one basis function for the \(x\)-direction and another for the \(y\)-direction
The most straightforward approach is to use the same basis functions for both directions. For example, with a Chebyshev basis
The two spatial directions have their own domain on the real line, and we use \(I_x = [a, b]\) for the \(x\)-direction and \( I_y = [c, d]\) for the \(y\)-direction. The domain is then the Cartesian product \(\Omega = I_x \times I_y\).
We can define two one-dimensional function spaces for the two directions as \(V_x = \text{span}\{\psi_i\}_{i=0}^M\) and \(V_y = \text{span}\{\varphi_i\}_{i=0}^N\). A two-dimensional function space can then be created as
The function space \(W\) is the tensor product of \(V_x\) and \(V_y\). The function space \(W\) has a basis that is formed as the tensor product of the basis for \(V_x\) and \(V_y\). The 2D basis function \(\Psi_{ij}\) is the tensor product of \(\psi_i(x)\) and \(\varphi_j(y)\)
This is also called the outer product, and sometimes also the dyadic product.
Note
The Kronecker product is a tensor product.
The tensor product is related to the Cartesian product. It may actually be viewed as the Cartesian product with multiplication. Consider the Cartesian product of the two sets \((1, 2, 3)\) and \((4, 5)\)
and compare with the tensor product
We see that the tensor product is in fact the Cartesian product with multiplication of the items in each set. The same applies to functions and function spaces. For \(V_x = \text{span}\{\psi_0(x), \psi_1(x)\}\) and \(V_y = \text{span}\{\varphi_0(y), \varphi_1(y)\}\) we have the Cartesian product of the bases
Similarly, the tensor product is:
This tensor product is the basis for \(W = V_x \otimes V_y\). Normally we would also shape the tensor product as a matrix. Here the first basis would be indexed as row and the second as column. This is the normal matrix indexing \(a_{ij}\), where \(i\) as row and \(j\) as column.
With matrix notation the tensor product can be understood as the matrix-matrix product of a column vector with a row vector:
For example, we can take the tensor product of the two bases \(\{1, x\}\) and \(\{1, y\}\) in Numpy:
y = sp.Symbol('y')
Vx = np.array([1, x])
Vy = np.array([1, y])
W = np.outer(Vx, Vy)
print(W)
[[1 y]
[x x*y]]
This is exactly
Lets try to approximate \(u(x,y) = \exp(-(x^2+2(y-0.5)^2))\), \((x, y) \in [-1, 1] \times [-1, 1]\) using Legendre polynomials and the Galerkin method. To this end we need the \(L^2(\Omega)\) inner product
Note that the generic first line is identical to the definition used for the 1D case (11), except that \(f\) and \(g\) now are functions of both \(x\) and \(y\).
For simplicity we use Legendre polynomials in both directions with the same dimension and space \(V_N = \text{span}\{P_i\}_{i=0}^N\) such that \(W = V_N \otimes V_N\). We now want to find \(u_N \in W\) such that
This means to compute
where the index set \(\mathcal{I}_N = \{0, 1, \ldots, N\}\) and \(\mathcal{I}_N^2 = \mathcal{I}_N \times \mathcal{I}_N\). The test function \(v\) is here the tensor product basis function \(P_m(x)P_n(y)\).
Note
Equation (35) provides \((N+1)^2\) equations for the \((N+1)^2\) unknown in \(\hat{U} = (\hat{u}_{ij})_{i,j=0}^N\).
Note that the unknown coefficients \(\hat{u}_{ij}\) are independent of space and we can simplify the double integrals by separating them into one integral for \(x\) and one for \(y\). We get
and we also write
We can thus write Eq. (35) as the linear algebra problem
In matrix form this is
which can be solved with the vec-trick as
However, since \(A\) is a diagonal matrix it is actually much easier to just avoid the vectorization and solve directly
Lets implement this and verify that the approximation is close to the exact solution. To this end we will now need to use the double integral functions dblquad. The rest is fairly straight forward:
import scipy.sparse as sparse
from scipy.integrate import dblquad
ue = sp.exp(-(x**2+2*(y-sp.S.Half)**2))
uh = lambda i, j: dblquad(sp.lambdify((x, y), ue*sp.legendre(i, x)*sp.legendre(j, y)), -1, 1, -1, 1, epsabs=1e-12)[0]
N = 20
uij = np.zeros((N+1, N+1))
for i in range(N+1):
for j in range(N+1):
uij[i, j] = uh(i, j)
A = sparse.diags([2/(2*np.arange(N+1)+1)], [0], (N+1, N+1))
A_inv = sparse.diags([(2*np.arange(N+1)+1)/2], [0], (N+1, N+1))
uhat_ij = A_inv @ uij @ A_inv
At this point we have the coefficients \(\hat{U}\). For comparison with the exact solution we will need to compute the solution in real space for a uniform mesh (meshgrid)
To this end we can make use of the Vandermonde matrix
and then (since the mesh is the same in the \(x\) and \(y\) directions) we can simply compute
Below we implement this and compute the \(l^2\) error norm using the exact solution.
xi = np.linspace(-1, 1, N+1)
V = np.polynomial.legendre.legvander(xi, N)
U = V @ uhat_ij @ V.T
xij, yij = np.meshgrid(xi, xi, indexing='ij', sparse=True)
ueij = sp.lambdify((x, y), ue)(xij, yij)
np.linalg.norm(U-ueij)*(2/N)
np.float64(7.519704442136279e-11)
Not bad at all. Lets try for a range of resolutions and plot the \(L^2\) error norm as a function of N
def convergence(N):
uij = np.zeros((N+1, N+1))
for i in range(N+1):
for j in range(N+1):
uij[i, j] = uh(i, j)
A = sparse.diags([2/(2*np.arange(N+1)+1)], [0], (N+1, N+1))
A_inv = sparse.diags([(2*np.arange(N+1)+1)/2], [0], (N+1, N+1))
uhat_ij = A_inv @ uij @ A_inv
xi = np.linspace(-1, 1, N+1)
V = np.polynomial.legendre.legvander(xi, N)
U = V @ uhat_ij @ V.T
xij, yij = np.meshgrid(xi, xi, indexing='ij', sparse=True)
ueij = sp.lambdify((x, y), ue)(xij, yij)
h = xi[1]-xi[0]
return np.sqrt(np.trapz(np.trapz((U-ueij)**2, dx=h), dx=h))
error = []
for n in range(4, 24, 2):
error.append(convergence(n))
/var/folders/ql/nly59sfj4dd3spkth6lk0kjw0000gn/T/ipykernel_55623/1651715329.py:15: DeprecationWarning: `trapz` is deprecated. Use `trapezoid` instead, or one of the numerical integration functions in `scipy.integrate`.
return np.sqrt(np.trapz(np.trapz((U-ueij)**2, dx=h), dx=h))
plt.figure(figsize=(5, 3))
plt.semilogy(np.arange(4, 24, 2), error);
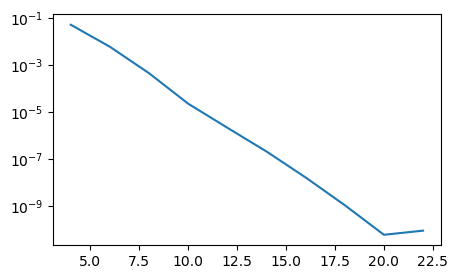
Note the extremely fast convergence. Doubling the number of points in each direction from \(N=10\) to \(N=20\) leads to an error that descreses with 5 decades! This is spectral accuracy. At \(N>20\) the solution does no longer improve because of roundoff errors and machine precision.
External software#
Two softwares that are using both Legendre and Chebyshev polynomials for global function approximation and to solve partial differential equations are:
Chebfun is mainly focused on Chebyshev polynomials and it is written in MATLAB. Shenfun is written in Python and focuses on solving partial differential equations via the Galerkin method.
Lets try to approximate
using Shenfun:
import shenfun as sf
ue = 1 / (1+16*x**2)
N = 20
V = sf.FunctionSpace(N+1, 'Legendre', domain=(-1, 1))
v = sf.TestFunction(V)
uh = (2*np.arange(N+1)+1)/2*sf.inner(ue, v, assemble='exact')
#uh = sf.project(ue, V) # or just this
The code should be fairly easy to follow. We simply create the function space V \(=\text{span}\{P_i\}_{i=0}^{N}\), then a test function v from this space and then we compute the Legendre coefficients using the formula in Eq. (32).
Note that there is a shortcut function project that projects \(u(x)\) to the chosen space and basis in one line of code.
We can plot the difference between the computed function and the exact function on a uniform mesh of length 1000:
xj = np.linspace(-1, 1, 1000)
plt.figure(figsize=(5, 3))
plt.plot(xj, uh(xj)-sp.lambdify(x, ue)(xj), 'r');
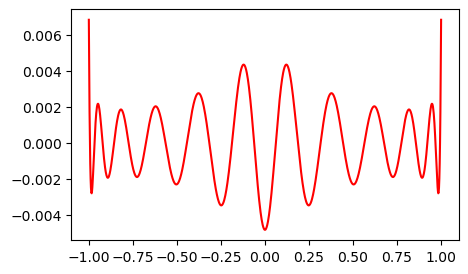
Notice how the error oscillates all throughout the domain. This is typical of spectral methods.
Lets do exactly the same for Chebyshev polynomials:
%%capture
C = sf.FunctionSpace(N+1, 'Chebyshev', domain=(-1, 1))
v = sf.TestFunction(C)
cj = np.ones(N+1)
cj[0] = 2
uhc = 2/(cj*np.pi)*sf.inner(ue, v, assemble='adaptive')
#uhc = sf.project(ue, C) # or just this
plt.figure(figsize=(5, 3))
plt.plot(xj, uhc(xj)- sp.lambdify(x, ue)(xj), 'r');
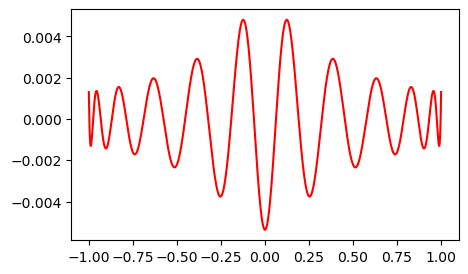
We see that the error is very similar to the Legendre, but slightly better towards the edges.
Two- and three-dimensional problems can also be solved using shenfun and tensor products. Lets try to approximate \(u(x, y) = \exp(-(x^2+2(y-0.5)^2)\) using Legendre polynomials in both directions and a tensor product function space.
ue = sp.exp(-(x**2+2*(y-sp.S.Half)**2))
N = 20
V = sf.FunctionSpace(N+1, 'Legendre')
W = sf.TensorProductSpace(sf.comm, (V, V))
uh = sf.project(ue, W)
xij, yij = W.local_mesh(True, kind='uniform')
plt.figure(figsize=(5, 3))
plt.contourf(xij, yij, uh.backward(mesh='uniform'));
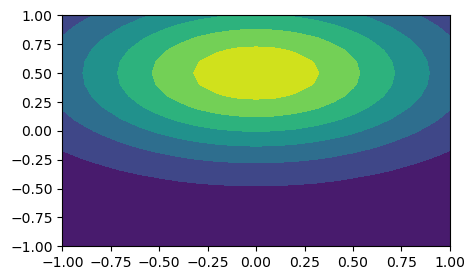
np.linalg.norm(uh.backward(mesh='uniform')-sp.lambdify((x, y), ue)(xij, yij))
np.float64(7.165905224632915e-10)
which is very similar to the already computed above without the use of Shenfun.
Fast Chebyshev transforms - (optional)#
Besides the closed form, the Chebyshev polynomials have another advantage over Legendre. The Chebyshev series can make use of fast Cosine transforms (similar to fast Fourier transforms) when evaluating a series for Chebyshev points.
Assume that the coefficients \((\hat{u}_j)_{j=0}^N\) are known and you want to evaluate this series on \(N+1\) points in physical space
Normally this can be computed as a matrix vector product
where the Vandermonde matrix \(\text{T} = (t_{ij})_{i,j=0}^N, t_{ij} = T_j(x_i)\) is of shape \((N+1) \times (N+1)\). The cost of this operation is \((2N+1)(N+1)\) floating point operations, and of leading order \(\mathcal{O}(N^2)\) .
Note
We compute the number of floating point operations for the matrix-vector product as follows: for each of the \(N+1\) rows in the matrix \(\text{T}\), we hold \(i\) constant and compute \(\sum_{j=0}^N t_{i, j} \cdot \hat{u}_j\). This requires \(N+1\) multiplications and \(N\) additions, so a total of \(2N+1\) flops. Since there are \(N+1\) rows in the matrix \(\text{T}\), this is repeated \(N+1\) times. The total cost is as such \((N+1)(2N+1)\) flops.
The matrix \(\text{T}\) can be computed using np.polynomial.chebyshev.chebvander, which can take any vector of points \((x_i)\), not necessarily of shape \(N+1\).
T = np.polynomial.chebyshev.chebvander(np.cos(np.arange(5)*np.pi/4), 5)
np.set_printoptions(precision=3, suppress=True)
T
array([[ 1. , 1. , 1. , 1. , 1. , 1. ],
[ 1. , 0.707, 0. , -0.707, -1. , -0.707],
[ 1. , 0. , -1. , -0. , 1. , 0. ],
[ 1. , -0.707, -0. , 0.707, -1. , 0.707],
[ 1. , -1. , 1. , -1. , 1. , -1. ]])
Now the column \(j\) in \(\text{T}\) represents the numbers \((T_j(x_i))_{i=0}^N\), that is, the basis function \(T_j\) evaluated on the mesh points \((x_i)_{i=0}^N\).
We can now evaluate (36) for any \(\boldsymbol{\hat{u}}\) simply as
uhat = np.array([0, 1, 0, 1, 0, 1])
T @ uhat
array([ 3. , -0.707, 0. , 0.707, -3. ])
Because of the choice of \(\hat{u} = (0, 1, 0, 1, 0, 1)\) we are now simply computing
where \(\boldsymbol{x} = (\cos(i \pi /5))_{i=0}^5\).
This matrix vector product is the straight forward approach that costs what it costs to do a matrix vector product. However, since the Vandermonde matrix \(\text{T}\) contains evaluated cosines, we can do better.
Lets evaluate the Chebyshev series at the Chebyshev points (25): \(x_i = \cos(i \pi /N), i \in 0, 1, \ldots, N\). Using the definition of Chebyshev polynomials (24) we get
for \(i = 0, 1, \ldots, N\).
The discrete cosine transform of type 1 is defined to transform the real numbers \(\boldsymbol{y} = (y_i)_{i=0}^N\) into \(\boldsymbol{Y}=(Y_i)_{i=0}^N\) such that
From the definition in Scipy we get that this \(DCT^{1}\) really computes
This is almost the same as (37), except that the sum has a factor 2 in front. But we can work around this. A minor reformulation leads to
where the right hand side now is exactly like in (37). So we need to compute
where \(I_m = ((-1)^{i})_{i=0}^N\). We can implement this as shown in the function evaluate_cheb_1 below
import scipy
def evaluate_cheb_1(uhat):
N = len(uhat)
uj = scipy.fft.dct(uhat, type=1)
uj += uhat[0]
uj[::2] += uhat[-1]
uj[1::2] -= uhat[-1]
uj *= 0.5
return uj
Lets make sure that this function computes the same as the matrix vector product defined above.
N = 1000
xi = np.cos(np.arange(N+1)*np.pi/N)
T = np.polynomial.chebyshev.chebvander(xi, N)
uhat = np.ones(N+1)
uj = T @ uhat
uj_fast = evaluate_cheb_1(uhat)
assert np.allclose(uj, uj_fast)
Since the assert is ok this means that the fast transform works.
Lets check the speed of the transforms
%timeit -n 10 uj_fast = evaluate_cheb_1(uhat)
The slowest run took 4.63 times longer than the fastest. This could mean that an intermediate result is being cached.
17.9 μs ± 14.8 μs per loop (mean ± std. dev. of 7 runs, 10 loops each)
%timeit -n 10 uj = T @ uhat
136 μs ± 13.3 μs per loop (mean ± std. dev. of 7 runs, 10 loops each)
Not a big difference, but this difference will become larger for larger arrays, because the cost of the fast transform is only \(\mathcal{O}(N \log_2 N)\). Also, the DCT from scipy is not very efficient. Faster versions are available from FFTW, wrapped for usage in Python through mpi4py-fft.
Weekly assignments#
Experiment with the Chebyshev Galerkin method and approximate the same global functions that was considered with Legendre and collocation last week:
\(u(x) = |x|, \quad x \in [-1, 1]\)
\(u(x) = \exp(\sin(x)), \quad x \in [0, 2]\)
\(u(x) = x^{10}, \quad x \in [0, 1]\)
\(u(x) = \exp(-(x-0.5)^2) - \exp(-0.25) \quad x \in [0, 1]\)
\(u(x) = J_0(x), \quad x \in [0, 100]\)
Approximate the function \(u(x, y) = \exp(-(x^2+2(y-0.5)^2))\) in the domain \(\Omega = [-1, 1]^2\) with Chebyshev polynomials in both \(x\) and \(y\)-directions. Try both to implement it yourself, as done for Legendre, and by using Shenfun.
